※Computational resources of protein cleavage, etc.:
Last updated: Feb. 26th, 2011
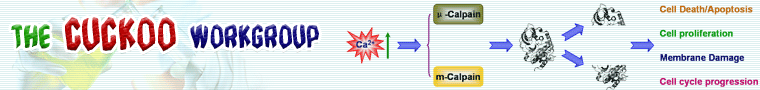
<1> Other resource of calpains and calpain-mediated cleavage.
1. SitePrediction: Predict the cleavage of substrates for proteinases including calpains(Verspurten, et al., 2009).
2. CaMPDB: a database on calpain, its substrates and calpastatin. A site-specific substrates prediction server was also provided.(duVerle, et al., 2010).
3. PoPS: a set of computational tools for investigating protease specificity. Users could input parameters such as score matrix to perform prediction for calpains.(Sarah, et al., 2005).
4. CutDB: a database focuses on the annotation of individual proteolytic events, both actual and predicted. The calpain mediated cleavage events were provided(Igarashi, et al., 2007).
<2> Other resource of protein cleavage
A. Proteases and Protease-mediated cleavage
1. MEROPS: a database for peptidases (also termed proteases, proteinases and proteolytic enzymes) and the proteins that inhibit them.(Rawlings, et al., 2004, Rawlings, et al., 2010).
2. Predict Putative Substrates of Proteases: A sequence and structure based method to predict putative substrates, functions and regulatory networks of endo proteases.(Venkatraman, et al., 2009).
3. The Degradome database: the peptidase database, which contains information on peptidases and inhibitors among organisms, substrate cleavage sites and indexes for expressed sequence tag libraries containing peptidases .(Quesada, et al., 2009).
4. Serine Proteases: a database of serine proteases.
5. PlantPIs: an interactive web resource on plant protease inhibitors.(De Leo, et al., 2009).
6. PeptideMass: cleaves a protein sequence from the UniProt Knowledgebase (Swiss-Prot and TrEMBL) or a user-entered protein sequence with a chosen enzyme, and computes the masses of the generated peptides.(Wilkins, et al., 1997).
B. Signal peptide cleavage sites
1. ChloroP: A web server predicts the presence of chloroplast transit peptides (cTP) in protein sequences and the location of potential cTP cleavage sites (Emanuelsson, et al., 1999).
2. PCLR: a method of predicting chloroplast localization of proteins in plant cells. The prediction algorithm was trained using principal component logistic regression with stepwise variable selection (Schein, et al., 2001).
3. LipoP: a web server produces predictions of lipoproteins and discriminates between lipoprotein signal peptides, other signal peptides and n-terminal membrane helices in Gram negative bacteria (Juncker, et al., 2003).
4. SpLip: a program that predicts lipoproteins in spirochetal genomes (Setubal, et al., 2006).
5. PRED-LIPO: Prediction of Lipoprotein and Secretory Signal Peptides in Gram-positive Bacteria with Hidden Markov Models (Bagos, et al., 2008).
6. MITOPROT: Prediction of mitochondrial targeting sequences (Claros and Vincens, 1996; Guda, et al., 2004).
7. PATS: PATS identifies amino acid sequences that are potentially targeted to the apicoplast matrix of Plasmodium falciparum. Note that secondary analysis of candidate sequences is required for confirmation (Waller, et al., 1998; Zuegge, et al., 2001).
8. PlasMit: Prediction of mitochondrial transit peptides in Plasmodium falciparum (Zuegge, et al., 2001).
9. Predotar: A prediction service for identifying putative N-terminal targeting sequences (Small, et al., 2004).
10. PTS1: Prediction of peroxisomal targeting signal 1 containing proteins from amino acid sequence (Neuberger, et al., 2003; Neuberger, et al., 2003).
11. PTS1Prowler: a predictor to screen several additional eukaryotic genomes to revise previously estimated numbers of peroxisomal proteins (Hawkins, et al., 2007).
12. PeroxisomeDB: a database for the peroxisomal proteome, functional genomics and disease (Schluter, et al., 2007).
13. PeroxiP: In silico prediction of the peroxisomal proteome in fungi, plants and animals (Emanuelsson, et al., 2003).
14. SignalP: a web server predicts the presence and location of signal peptide cleavage sites in amino acid sequences from different organisms: Gram-positive prokaryotes, Gram-negative prokaryotes, and eukaryotes. The method incorporates a prediction of cleavage sites and a signal peptide/non-signal peptide prediction based on a combination of several artificial neural networks and hidden Markov models (Nielsen, et al., 1999; Bendtsen, et al., 2004; Emanuelsson, et al., 2007; Nielsen, et al., 1997; Menne, et al., 2000).
15. TatP 1.0: a web server predicts the presence and location of Twin-arginine signal peptide cleavage sites in bacteria. The method incorporates a prediction of cleavage sites and a signal peptide/non-signal peptide prediction based on a combination of two artificial neural networks. A postfiltering of the output based on regular expressions is possible (Bendtsen, et al., 2005).
16. ProP 1.0: a web server predicts arginine and lysine propeptide cleavage sites in eukaryotic protein sequences using an ensemble of neural networks. Furin-specific prediction is the default. It is also possible to perform a general proprotein convertase (PC) prediction (Duckert, et al., 2004).
17. TargetP 1.1: a web server predicts the subcellular location of eukaryotic proteins. The location assignment is based on the predicted presence of any of the N-terminal presequences: chloroplast transit peptide (cTP), mitochondrial targeting peptide (mTP) or secretory pathway signal peptide (SP) (Nielsen, et al., 1997; Emanuelsson, et al., 2000).
18. MultiLoc: prediction of protein subcellular localization using N-terminal targeting sequences, sequence motifs, and amino acid composition (Hoglund, et al., 2006).
19. NetCorona 1.0: a web server predicts coronavirus 3C-like proteinase (or protease) cleavage sites using artificial neural networks on amino acid sequences. Every potential site is scored and a list is compiled in addition to a graphical representation. Refer to publication for more detailed information and performance values (Kiemer, et al., 2004).
20. NetPicoRNA 1.0: a web server produces neural network predictions of cleavage sites of picornaviral proteases. The method is described in detail in the reference article mentioned below (see CITATIONS at the bottom of this page) (Blom, et al., 1996).
21. SecretomeP 2.0: a web server produces ab initio predictions of non-classical i.e. not signal peptide triggered protein secretion. The method queries a large number of other feature prediction servers to obtain information on various post-translational and localizational aspects of the protein, which are integrated into the final secretion prediction (Bendtsen, et al., 2004; Bendtsen, et al., 2005).
22. NetNES 1.1: a web server predicts leucine-rich nuclear export signals (NES) in eukaryotic proteins using a combination of neural networks and hidden Markov models (la Cour, et al., 2004).
23. PeptideCutter: a web server predicts potential cleavage sites cleaved by proteases or chemicals in a given protein sequence. PeptideCutter returns the query sequence with the possible cleavage sites mapped on it and /or a table of cleavage site positions (Wilkins, et al., 1999).
24. SPOCTOPUS: a combined predictor of signal peptides and membrane protein topology (Viklund, et al., 2008).
25. HECTAR: a method to predict subcellular targeting in heterokonts (Gschloessl, et al., 2008).
26. Signal-BLAST: High-performance signal peptide prediction based on sequence alignment techniques (Frank and Sippl, 2008).
27. PredSL: a tool for the N-terminal sequence-based prediction of protein subcellular localization (Petsalaki, et al., 2006).
28. iPSORT: Extensive feature detection of N-terminal protein sorting signals (Bannai, et al., 2002).
29. Phobius in Sweden; Phobius in UK: A combined transmembrane topology and signal peptide predictor (Kall, et al., 2004; Kall, et al., 2007).
30. PrediSi: prediction of signal peptides and their cleavage positions (Hiller, et al., 2004).
31. RPSP: Prediction of signal peptides in protein sequences by neural networks (Plewczynski, et al., 2008).
32. Signal-3L: A 3-layer approach for predicting signal peptides (Shen and Chou, 2007).
33. Signal-CF: a subsite-coupled and window-fusing approach for predicting signal peptides (Chou and Shen, 2007).
34. HIVcleave: Predicting HIV protease cleavage sites in proteins (Chou, 1996; Shen and Chou, 2008).
35. TISs-ST:
a web server to evaluate polymorphic translation
initiation sites and their reflections on
the secretory targets (Vicentini
and Menossi, 2007).
C. Caspase substrates cleavage
1. CASVM: Server for SVM Prediction of Caspase Substrates Cleavage Sites (Wee, et al., 2006; Wee, et al., 2007).
2. GraBCas: A bioinformatics tool for score-based prediction of Caspase- and Granzyme B-cleavage sites in protein sequences (Backes, et al., 2005).
3. PEPS: is a tool for the prediction of caspase caspase cleavage sites. It is based on the consensus motifs for caspase substrates. Software can be obtained upon request from author (Lohmuller, et al., 2003).
4. CaSPredictor:
is a software tool for the prediction of caspase
cleavage sites which utilizes position-specific
scoring matrices together with indices based
on neighboring PEST sequences. Software can
be obtained upon request from author (Garay-Malpartida, et al., 2005).
D. Proteasomal cleavage
1. NetChop 3.0: a web server produces neural network predictions for cleavage sites of the human proteasome (Nielsen, et al., 2005).
2. PepCleave: Precise score for the prediction of peptides cleaved by the proteasome (Ginodi, et al., 2008).
3. MAPPP: MHC class I antigenic peptide processing prediction (Hakenberg, et al., 2003).
4. Paproc: a prediction tool for cleavages by human and yeast 20S proteasomes, based on experimental cleavage data (Nussbaum, et al., 2001).
5. Pcleavage: an SVM based method for prediction of constitutive proteasome and immunoproteasome cleavage sites in antigenic sequences (Vicentini and Menossi, 2007).