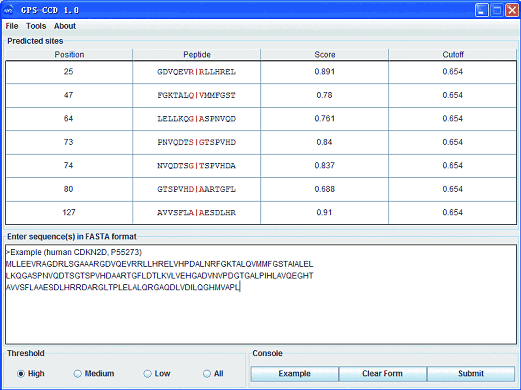
※ GPS-CCD INTRODUCTION:
Calpains constitute an important family of the Ca2+-dependent cysteine proteases, which contain a nucleophilic cysteine in the catalytically active site (Goll et al, 2003; Franco et al, 2005; Zatz et al, 2005; Liu et al, 2008). Calpains are widely expressed in mammalians and conserved across eukaryotes (Futai et al, 2001; Croall et al, 2007). For instance, in budding yeast, at least one calpain-like protease, Rim13/Cpl1, has been identified, although its functions are still elusive (Hayashi et al, 2001). In humans, there are over 14 distinct members of the calpain superfamily, some of which are tissue specific. Calpain 1 (μ-calpain, micromolar Ca2+-requiring) and Calpain 2 (m-calpain, millimolar Ca2+-requiring) are ubiquitously expressed and well characterized isoforms (Huang et al, 2005). Through spatial and temporal cleavage of a variety of substrates to change their conformation, function and stability (Glading et al, 2002), Ca2+-activated calpains play an important role in numerous biological processes, including the regulation of gene expression, signal transduction, cell death/apoptosis, remodeling cytoskeletal attachments during cell fusion/motility and cell cycle progression (Squier et al, 1999; Tan et al, 2006; Yousefi et al, 2006). Moreover, calpain aberrancies are frequently implicated in a variety of diseases and cancers (Arrington et al, 2006; Williams et al, 2008). Although many studies have tried to dissect the regulatory roles and molecular mechanisms of calpain-dependent cleavage, in fact our understanding of calpain is still fragmentary.
In this work, we collected 368 experimentally verified calpain cleavage sites in 130 proteins. With a previously developed algorithm of GPS (Group-based Prediction System)(Xue et al, 2008), we developed a novel software package of GPS-CCD (Calpain Cleavage Detector) for the prediction of calpain cleavage sites. The leave-one-out validation and 4-, 6-, 8-, 10-fold cross-validations were performed to evaluate the performance of the prediction system. By comparison, the GPS 2.0 algorithm was employed for its outstanding prediction performance, with an accuracy 89.98%, sensitivity 60.87% and specificity 90.07%. Furthermore, there are many proteins experimentally identified as calpain substrates for which the exact cleavage sites have not been verified, and we collected 196 such proteins from PubMed. As an application, we predicted potential calpain cleavage sites for these targets. These prediction results might be a useful resource for further experimental investigation. Finally, the online service and local packages of GPS-CCD 1.0 were implemented in JAVA 1.5 (J2SE 5.0) and are freely available for academic researchers at: http://ccd.biocuckoo.org/.
GPS-CCD 1.0 User Interface
For publication of results please cite the following article:  GPS-CCD: GPS-CCD: A novel computational program for the prediction of calpain cleavage sites
GPS-CCD: GPS-CCD: A novel computational program for the prediction of calpain cleavage sites
Zexian Liu, Jun Cao, Xinjiao Gao, Qian Ma,Jian Ren and Yu Xue. PLoS ONE, 2011, 6(4): e19001 |